Principal Component Analysis
Function Overview
Calculate the eigenvalue, covariance, correlation coefficient and positive selection of each data.
Usage
Click Spectrum > Principal Component Analysis.
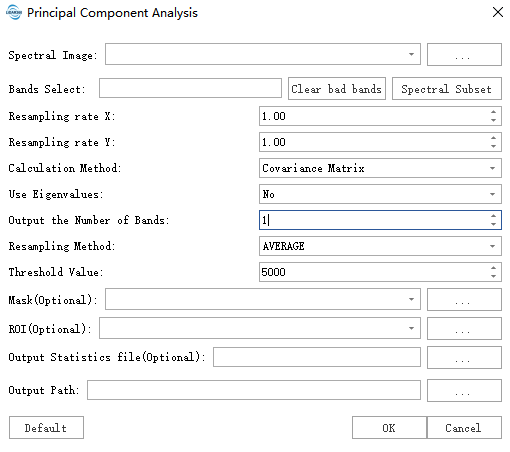
Click the Spectrum Subset button to select the band of the hyperspectral image.

Parameter Setting
- Input hyperspectral image data: Input the hyperspectral image data file. Data should be pre-processed, such as radiation calibration and atmospheric correction.
- Band selection: Select the band of the hyperspectral image. For PCA function, please click Clear the bad band before using it.
- Resampling rate X: Used to set the ratio of resampling. The smaller the value, the fewer the number of cells involved in the calculation, the faster the calculation speed, the range is 0-1.
- Resampling rate Y: Used to set the ratio of resampling. The smaller the value, the fewer the number of cells involved in the calculation, the faster the calculation speed, the range is 0-1.
- Calculation method: Covariance matrix: suitable for situations where data differences between bands are small. Correlation coefficient matrix: suitable for situations where data differences between bands are large.
- Resampling Method: Specifies the resampling technique that will be used to build the pyramids.
- Threshold Value: The processing threshold (default value is 5000) is used to adjust the analysis precision. A higher value results in greater precision but slower processing speed.
- ROI: Only the minimum external rectangle in the ROI area is calculated after selection.
- Mask: Only the effective area of the mask is calculated after selection.
- Output MNF file: Output MNF statistics.
- Output path: The generated tif path.